UMAP Embeddings¶
Uniform Manifold Approximation and Projection (UMAP) is a dimensinoal reduction technique. See the full documentation here.
from os import environ
from pathlib import Path
import numpy as np
import umap
import umap.plot
import GSForge as gsf
import holoviews as hv
import matplotlib.patches as mpatches
import matplotlib.pyplot as plt
hv.extension("bokeh")
OSF_PATH = Path(environ.get("GSFORGE_DEMO_DATA", default="~/GSForge_demo_data/osfstorage/oryza_sativa")).expanduser()
NORMED_GEM_PATH = OSF_PATH.joinpath("AnnotatedGEMs", "oryza_sativa_hisat2_normed.nc")
agem = gsf.AnnotatedGEM(NORMED_GEM_PATH)
agem
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
~/virtualenv/python3.7.1/lib/python3.7/site-packages/xarray/backends/file_manager.py in _acquire_with_cache_info(self, needs_lock)
198 try:
--> 199 file = self._cache[self._key]
200 except KeyError:
~/virtualenv/python3.7.1/lib/python3.7/site-packages/xarray/backends/lru_cache.py in __getitem__(self, key)
52 with self._lock:
---> 53 value = self._cache[key]
54 self._cache.move_to_end(key)
KeyError: [<class 'netCDF4._netCDF4.Dataset'>, ('/home/travis/GSForge_demo_data/AnnotatedGEMs/oryza_sativa_hisat2_normed.nc',), 'r', (('clobber', True), ('diskless', False), ('format', 'NETCDF4'), ('persist', False))]
During handling of the above exception, another exception occurred:
FileNotFoundError Traceback (most recent call last)
/tmp/ipykernel_5750/3720900272.py in <module>
----> 1 agem = gsf.AnnotatedGEM(NORMED_GEM_PATH)
2 agem
~/virtualenv/python3.7.1/lib/python3.7/site-packages/GSForge/models/_AnnotatedGEM.py in __init__(self, *args, **params)
65 logger.debug('Initializing a new gsforge.AnnotatedGEM object...')
66 if args:
---> 67 params = self.__annotated_gem_dispatch(*args, **params)
68 super().__init__(**params)
69 logger.debug('AnnotatedGEM initialization complete.')
~/virtualenv/python3.7.1/lib/python3.7/site-packages/GSForge/_singledispatchmethod.py in _method(*args, **kwargs)
71 def _method(*args, **kwargs):
72 method = self.dispatcher.dispatch(args[0].__class__)
---> 73 return method.__get__(obj, cls)(*args, **kwargs)
74
75 _method.__isabstractmethod__ = self.__isabstractmethod__
~/virtualenv/python3.7.1/lib/python3.7/site-packages/GSForge/models/_AnnotatedGEM.py in _parse_netcdf_path(cls, netcdf_path, **params)
172 A parsed parameter dictionary.
173 """
--> 174 params = cls._parse_xarray_dataset(xr.open_dataset(netcdf_path), **params)
175 return {"data": xr.open_dataset(netcdf_path), **params}
176
~/virtualenv/python3.7.1/lib/python3.7/site-packages/xarray/backends/api.py in open_dataset(filename_or_obj, engine, chunks, cache, decode_cf, mask_and_scale, decode_times, decode_timedelta, use_cftime, concat_characters, decode_coords, drop_variables, backend_kwargs, *args, **kwargs)
499 drop_variables=drop_variables,
500 **decoders,
--> 501 **kwargs,
502 )
503 ds = _dataset_from_backend_dataset(
~/virtualenv/python3.7.1/lib/python3.7/site-packages/xarray/backends/netCDF4_.py in open_dataset(self, filename_or_obj, mask_and_scale, decode_times, concat_characters, decode_coords, drop_variables, use_cftime, decode_timedelta, group, mode, format, clobber, diskless, persist, lock, autoclose)
558 persist=persist,
559 lock=lock,
--> 560 autoclose=autoclose,
561 )
562
~/virtualenv/python3.7.1/lib/python3.7/site-packages/xarray/backends/netCDF4_.py in open(cls, filename, mode, format, group, clobber, diskless, persist, lock, lock_maker, autoclose)
378 netCDF4.Dataset, filename, mode=mode, kwargs=kwargs
379 )
--> 380 return cls(manager, group=group, mode=mode, lock=lock, autoclose=autoclose)
381
382 def _acquire(self, needs_lock=True):
~/virtualenv/python3.7.1/lib/python3.7/site-packages/xarray/backends/netCDF4_.py in __init__(self, manager, group, mode, lock, autoclose)
326 self._group = group
327 self._mode = mode
--> 328 self.format = self.ds.data_model
329 self._filename = self.ds.filepath()
330 self.is_remote = is_remote_uri(self._filename)
~/virtualenv/python3.7.1/lib/python3.7/site-packages/xarray/backends/netCDF4_.py in ds(self)
387 @property
388 def ds(self):
--> 389 return self._acquire()
390
391 def open_store_variable(self, name, var):
~/virtualenv/python3.7.1/lib/python3.7/site-packages/xarray/backends/netCDF4_.py in _acquire(self, needs_lock)
381
382 def _acquire(self, needs_lock=True):
--> 383 with self._manager.acquire_context(needs_lock) as root:
384 ds = _nc4_require_group(root, self._group, self._mode)
385 return ds
/opt/python/3.7.1/lib/python3.7/contextlib.py in __enter__(self)
110 del self.args, self.kwds, self.func
111 try:
--> 112 return next(self.gen)
113 except StopIteration:
114 raise RuntimeError("generator didn't yield") from None
~/virtualenv/python3.7.1/lib/python3.7/site-packages/xarray/backends/file_manager.py in acquire_context(self, needs_lock)
185 def acquire_context(self, needs_lock=True):
186 """Context manager for acquiring a file."""
--> 187 file, cached = self._acquire_with_cache_info(needs_lock)
188 try:
189 yield file
~/virtualenv/python3.7.1/lib/python3.7/site-packages/xarray/backends/file_manager.py in _acquire_with_cache_info(self, needs_lock)
203 kwargs = kwargs.copy()
204 kwargs["mode"] = self._mode
--> 205 file = self._opener(*self._args, **kwargs)
206 if self._mode == "w":
207 # ensure file doesn't get overriden when opened again
src/netCDF4/_netCDF4.pyx in netCDF4._netCDF4.Dataset.__init__()
src/netCDF4/_netCDF4.pyx in netCDF4._netCDF4._ensure_nc_success()
FileNotFoundError: [Errno 2] No such file or directory: b'/home/travis/GSForge_demo_data/AnnotatedGEMs/oryza_sativa_hisat2_normed.nc'
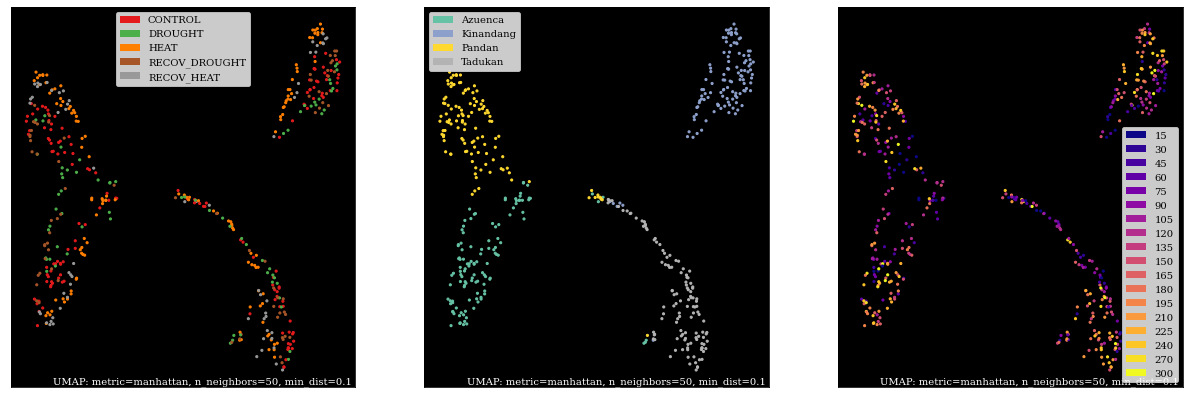
Plotting with umap.plot¶
By far the easiest method.
counts, labels = gsf.get_gem_data(agem, annotation_variables=['treatment', 'genotype', 'time'],
count_transform=lambda c: np.log2(c + 0.25))
mapper = umap.UMAP(random_state=42, metric='manhattan', n_neighbors=50).fit(counts.values)
fig, axes = plt.subplots(1, 3, figsize=(21, 7))
umap.plot.points(mapper, labels=labels['treatment'], background='black', ax=axes[0], color_key_cmap='Set1');
umap.plot.points(mapper, labels=labels['genotype'], background='black', ax=axes[1], color_key_cmap='Set2');
umap.plot.points(mapper, labels=labels['time'], background='black', ax=axes[2], color_key_cmap='plasma');
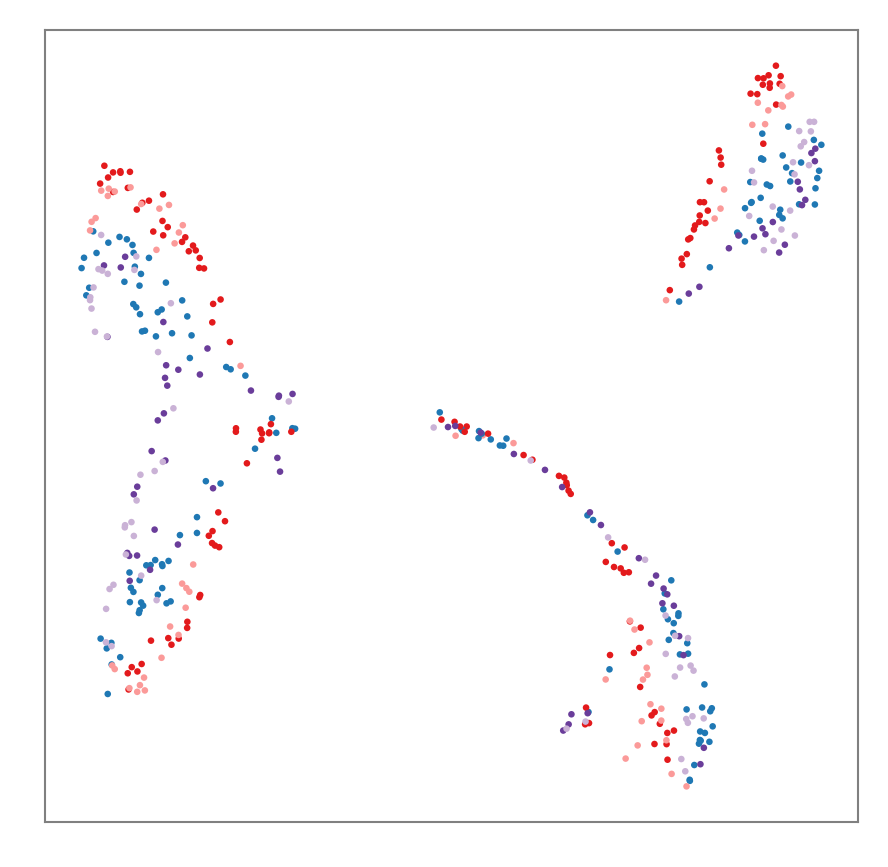
Plotting Manually¶
For posters, papers and other occasions that may have specific formatting requirements you will want to access the embedding data directly and construct your plot to said requirements.
# Set up custom colors.
treatment_cmap = {
'CONTROL': '#1f78b4',
'HEAT': '#e31a1c',
'RECOV_HEAT': '#fb9a99',
'DROUGHT': '#6a3d9a',
'RECOV_DROUGHT': '#cab2d6',
}
treatment_colors = labels['treatment'].to_series().map(treatment_cmap).values
# Declare fonts and sizes per your requirements.
plt.rcParams.update({'font.size': 10, 'font.family': 'serif'})
# Create the figure at the right size and resolution.
fig_inches = 3.5
fig_dpi = 300 # 300 is a common DPI requirement.
# Construct the figure.
fig, ax = plt.subplots(figsize=(fig_inches, fig_inches), dpi=fig_dpi)
# Fuss with the ticks. The numerical output of UMAP is not so important here.
ax.xaxis.set_visible(False)
ax.yaxis.set_visible(False)
for spine in ax.spines.values():
spine.set_edgecolor('grey')
spine.set_linewidth(0.5)
# Add the scatter points.
ax.scatter(mapper.embedding_[:, 0], mapper.embedding_[:, 1], s=10, marker='.', linewidths=0, c=treatment_colors);
handles = [mpatches.Patch(color=c, label=k) for k, c in treatment_cmap.items()]
fig, axes = plt.subplots(dpi=fig_dpi, figsize=(3, 0.75))
axes.axis('off')
axes.legend(handles=handles, ncol=2);
